You are here: Foswiki>Homepages Web>HomepageJacekOtwinowski>TeachingWorkshop>Dielectrons (2007-07-05, JacekOtwinowski)Edit Attach
- Dielectron Signal and Combinatorial Background (CB)
- CB reconstruction
- CB description in HADES
- CB reduction in HADES
- How to run e+e- pair analysis in the HYDRA framework
- Example of programs
- Exercises (HTree with HPair category with C+C@1AGeV events)
- Exercises (control HPairFilter ntuple with C+C@1GeV events)
Dielectron Signal and Combinatorial Background (CB)
All single electrons and positrons reconstructed in one event are combined into unlike-sign (e+e-) and like-sign (e+e+, e-e-) pairs in such a way that multiple combinations of the same particle are omitted. Besides the signal pairs arising from Dalitz and direct decays of light (M < 1.1 GeV/c^2) mesons and baryons, there are a lot of fake e+e- pairs which come from wrong combinations of e+ and e- tracks. These fake e+e- pairs contribute to huge combinatorial background (CB) which must be reduced and properly reconstructed. It should be noted that uncorrelated and correlated fake e+e- pairs contribute to CB background. The e+e- signal can be written asSignal = N(e+e-)-CB where N(e+e-) is a number of all unlike-sign pairs.
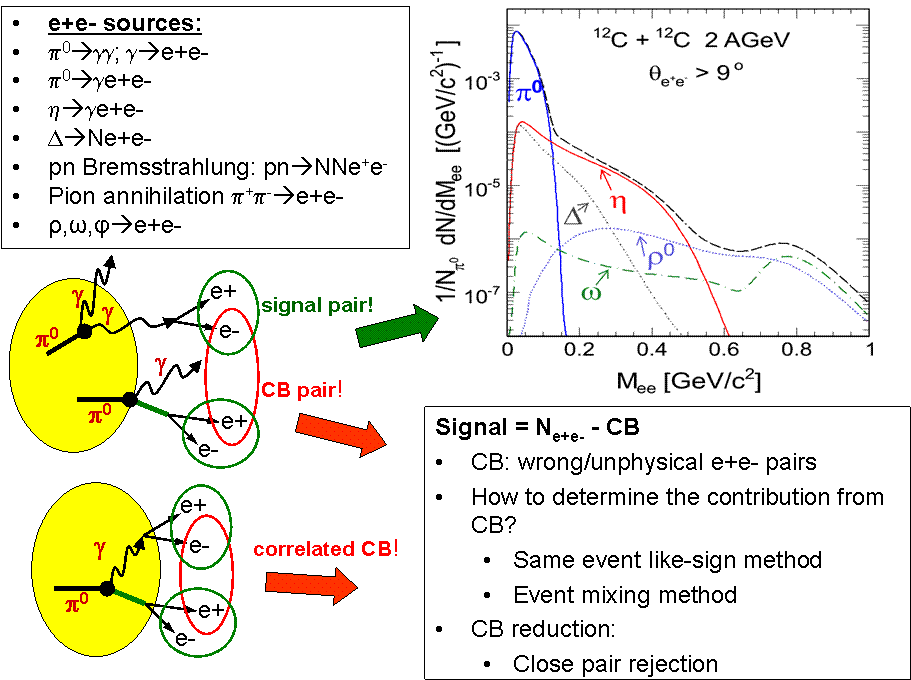
CB reconstruction
There are two methods of dielectron combinatorial background (CB) reconstruction which are used in HADES. The first method is called "Same Event Like-Sign" (SE) while the second method is called "Event Mixing" (EM). The SE method uses like-sign (e+e+, e-e-) pairs from the same event. It can be shown that the geometric mean value of the number of like-sign pairs (2*sqrt(N(e+e+)*N(e-e-))) reconstructed in the same event properly describe the CB. This formula is valid for Eff(e+e+)=Eff(e-e-)=Eff(e+e-) where Eff(ee) denotes efficiency of pair reconstruction. On the other hand the EM method calculates CB by combining e+ and e- coming from two different events into unlike-sign pairs. Thus, the CB obtained by the EM method is purly uncorrelated.
Both methods have advantages and disadvantages. The SE methods properly descibes correlated and uncorrelated CB (shape and yield), hovewer has statistical limitations (small number of like-sign pairs in one event). On the other side, The EM method does not have statistical limitations (arbitral number of events for mixing) but properly descibes only shape of uncorrelated CB.
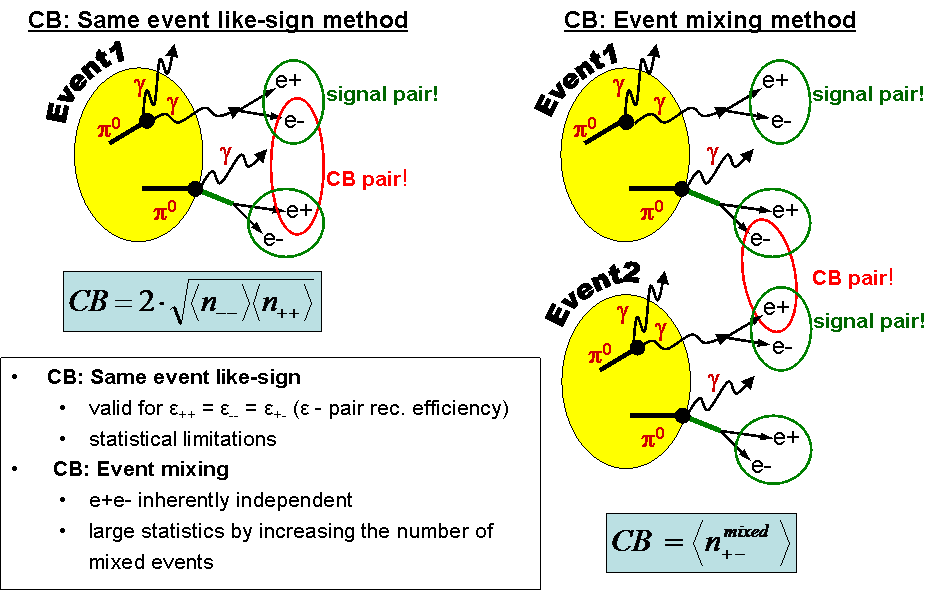
CB description in HADES
The CB reconstruction in HADES is done by using combination of SE and EM methods. For small e+e- invariant masses (M<150 MeV/c^2) the CB is reconstructed by means of SE method, while for the larger e+e- invariant masses the EM method is chosen.The CB obtained by the EM method is normalised in such a way to get the same integrated yiel as SE CB in the selected range. Such procedure gives us proper CB description for small invariant masses (correlated and uncorrelated CB) as well as for large invariant masses (small pair statistics, uncorrelated CB).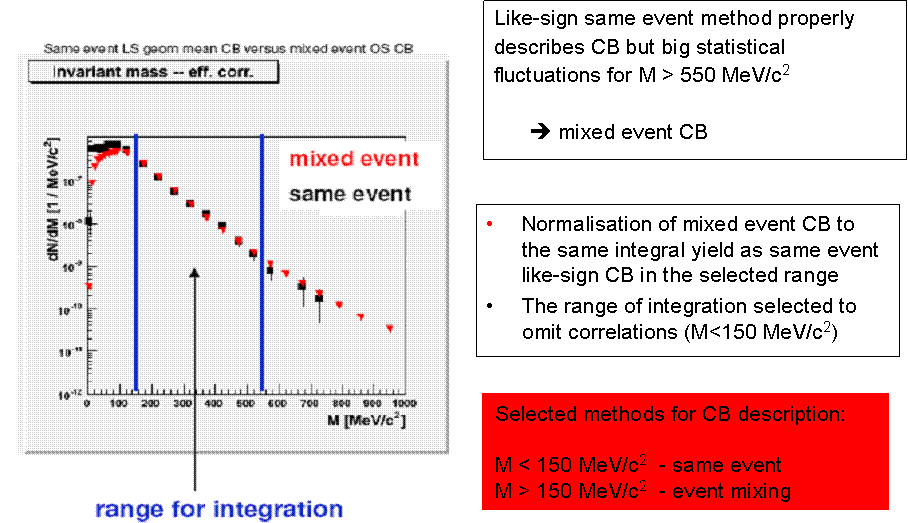
CB reduction in HADES
In order to reduce a huge CB the several pair selection cuts are implemented in the Hydra framework. By default the double hit and close pair rejecton cuts are used to reduce CB background. The first of them rejects all tracks which share the same hit in any of detectors of the HADES system. The second rejects all tracks which have close partner (opening angle < 9 deg.). In addition, the pairs with small opening angle (opening angle < 9 deg.) are removed recursively from the sample. In the pair analysis "direct" and "recursive" cuts are used. The "direct" cuts reject all pairs with small openinig angle, while "recursive" reject also both tracks belonging to this pair to omit further combinations. It means that the other pairs which contain at least one such track will be also rejected from the sample.Topology of unlike-sign pairs
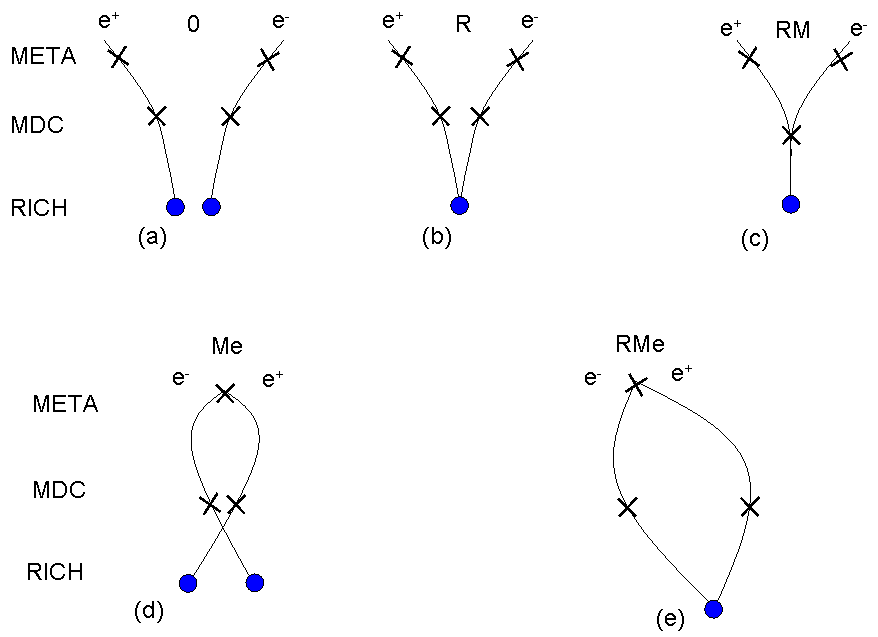
Topology of like-sign pairs
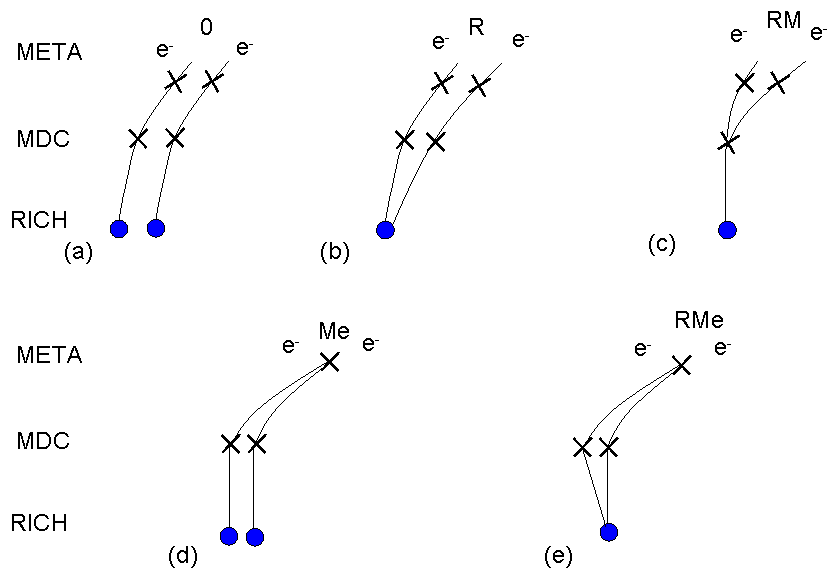
Close pairs rejection

How to run e+e- pair analysis in the HYDRA framework
TheHPairFiller and HPairFilter reconstructors are used in HYDRA framework to run e+e- analysis. The HPairFiller takes identified particles from HPidParticle/HPidParticleSim data category and creates all possible (unique) pairs in the event. However, only lepton-lepton and lepton-hadron pairs in which leptons have associated Rich rings are stored in the ouput HPair/HPairSim data category. The HPairFilter reconstructor filters pairs from HPair/HPairSim category by applying cuts retrived from HPairCutPar parameter container. If the pair doesn't pass the cut, the HPairFilter reconstructor sets (for this pair) the bit value to 0 in the array of bits, in the place corresponding to the cut number.
Each of the reconstructors can also create the output control ntuples (optional) with useful information. The HPairFiller control ntuple contains all possible pairs (hadron-hadron, hadron-lepton, and lepton-lepton) while the HPairFilter control ntuple contains pairs stored in HPair/HPairSim data category.
At the moment the HPairCutPar parameter container contains following cuts:
################################################################################
# Class: HPairCutPar
# Context: PairDefault
################################################################################
[PairCutParameters]
author: Oracle
description: Valid for: 01-MAR-2004 00:00:01 - 01-JAN-4000 00:00:00 \
Status At: 27-JUN-2007 10:41:41
//------------------------------------------------------------------------------
PairOpeningAngle: f 9.000
PairOpeningAngleFlag: i 1
PairOpeningAngleRecFlag: i 1
DoubleDetectorHitFlag: i 1
DoubleDetectorHitRecFlag: i 1
ZRichExit: f 27.98
PairDist: f 5.
ZRichBulb: f 1.0
ZTargetShift: f -13.5
RFlanch: f 32.0
XBeamCenter: f -2.5
YBeamCenter: f -4.0
BeamRadius: f 6.0
PairDistFlag: i 0
PairVertexFlag: i 0
################################################################################
By default the opening angle and double hit conditions are checked for the pair. The rest of parameters are used to calculate the space distance of tracks of the pair partners or pair production vertex.
In order to run the pair analysis in the HYDRA framework the following lines of code have to be added to the analysis program (default settings for "real" events):
// create HPairFiller reconstructor (must be added later to task set)
HPairFiller *pPairFiller=new HPairFiller("HPairFiller","HPairFiller");
// create HPairFilter reconstructor (must be added later to task set)
HPairFilter *pPairFilter=new HPairFilter("HPairFilter","HPairFilter");
// create output pPairFilter control ntuple (optional)
pPairFilter->setQAFileName("ntupleFiltFileName.root");
// add HPairFiller and HPairFilter reconstructors/tasks to the master task set
HTaskSet *pTaskSet=gHades->getTaskSet("real");
pTaskSet->add(pPairFiller);
pTaskSet->add(pPairFilter);
Example of programs
program to run pair analysis for "real" or "simulation" events Exercises (HTree with HPair category with C+C@1AGeV events)
- Draw
HPairdata members using TBrowser- Open pair code documentation http://hades-wiki.gsi.de/cgi-bin/view/SimAna/PairCodeDocumentation and find
HPairdescription - Open PostDST file (
root -l /d/hades06/data/jacek/aug04/be04253052729_dst_gen3_dil.root) - Create TBrowser object (e.g.
TBrowser a) - Go to
ROOT Filesand findHPair.fDatabranch in theTtree - Draw the following variables:
HPair.fData.nPid1,HPair.fData.nPid2,HPair.fData.fInvMass,HPair.fData.fOpang
- Open pair code documentation http://hades-wiki.gsi.de/cgi-bin/view/SimAna/PairCodeDocumentation and find
- Draw
HPair.fData.fInvMassfor unlike-sign and like-sign pairs (e.g.T.Draw("HPair.fData.fInvMass","(HPair.fData.nPid1==2&&HPair.fData.nPid2==3)||(HPair.fData.nPid1==3&&HPair.fData.nPid2==2)"))
Exercises (control HPairFilter ntuple with C+C@1GeV events)
- Draw invariant mass, opening angle, transverse momentum and rapidity plots for unlike-sign (e-e+) and like-sign (e-e-,e+e+) pairs
- Open http://hades-wiki.gsi.de/cgi-bin/view/SimAna/PairCodeDocumentation and find descritpion of pair ntuple
- Open the
HPairFilterpair ntuple (root -l /d/hades06/data/jacek/aug04/pairs_FULL_GOOD.root) - Create TBrowser object and open
PairsFilterntuple - Draw
pid1andpid2variables - Draw invariant mass (
invmass), opening angle (opang), transverse momentum (pt), rapidity (rap) distributions for unlike-sign and like-sign pairs e.g.:-
PairsFilter->Draw("invmass>>m_epem(100,0.,1000.)","(pid1==2&&pid2==3)||(pid1==3&&pid2==2)") -
PairsFilter->Draw("opang>>oa_epem(100,0.,180.)","(pid1==2&&pid2==3)||(pid1==3&&pid2==2)") -
PairsFilter->Draw("pt>>pt_epem(100,0.,1000.)","(pid1==2&&pid2==3)||(pid1==3&&pid2==2)") -
PairsFilter->Draw("rap>>rap_epem(100,0.,2.)","(pid1==2&&pid2==3)||(pid1==3&&pid2==2)")
-
- Draw
invariant massspactra for unlike-sign and like-sign pairs as the function of direct and recursive cuts (comparison)- Available cuts:
- isGoodOpang - direct opening angle cut flag
- isNotDoubleHit - direct double hit cut flag
- isCutNb - direct and recursive cut flags
isCutNb: info if pair was cut or not const UInt_t HPairCutPar::NOCUT = 0; const UInt_t HPairCutPar::DIRECTCUT = 1; const UInt_t HPairCutPar::OPANG = 2; // track removing const UInt_t HPairCutPar::DOUBLEHIT = 3; // track removing const UInt_t HPairCutPar::OPANGDOUBLEHIT = 10; // track removing - track environment conditions (angular distance to the closest neighbour)
- Draw
isCutNbvariable ("double hit and opening angle recursive cuts")-
PairsFilter->Draw("isCutNb")
-
- Draw track environment varibles:
angletoclosestfittedlep1,angletoclosestnonfittedlep1,angletoclosestfittedhad1,angletoclosestnonfittedhad1(e.g.PairsFilter->Draw("angletoclosestfittedlep1>>afl(100,0,20)")) - Draw
invariant massspectra for unlike-sign and like-sign pairs after all direct cuts (e.g.PairsFilter->Draw("invmass>>m_epem_dc(100,0.,1000.)","((pid1==2&&pid2==3)||(pid1==3&&pid2==2)) && isGoodOpang==1 && isNotDoubleHit==1") - Draw
invariant massspactra for unlike-sign and like-sign pairs after all recursive cuts (e.g.PairsFilter->Draw("invmass>>m_epem_rc(100,0.,1000.)","((pid1==2&&pid2==3)||(pid1==3&&pid2==2)) && isCutNb==0")) - Draw
m_epem_dcandm_epem_rchistograms in one canvas for comparison - Draw
m_emem_dcandm_emem_rchistograms in one canvas for comparison - Draw
invariant massspectrum for unlike-sign pairs after all recursive cuts and environment conditions (e.g.isCutNb==0 && angletoclosestnonfittedlep1>9. && angletoclosestnonfittedlep2>9. && angletoclosestfittedlep1>9. && angletoclosestfittedlep2>9.)
- Available cuts:
- Calculate dielectron signal
S = N(e-e+)-CB (CB = N(e+e+)+N(e-e-))(after recursive cuts)- Use ROOT
Clone()function to clonem_epem_rc,m_epep_rcandm_emem_rchistograms (e.g.TH1F *m_epem_rc_clone = m_epem_rc->Clone()) - Calculate combinatorial background CB (
m_epep_rc_clone->Add(m_emem_rc_clone,1)) - Calculate e+e- signal (
m_epem_rc_clone->Add(m_epep_rc_clone,-1)) - Draw e+e- pairs, e+e- signal and CB in the same canvas (
m_epem_rc->Draw();m_epem_rc_clone->Draw("same");m_epep_rc_clone->Draw("same"))
- Use ROOT
| I | Attachment | Action | Size | Date | Who | Comment |
|---|---|---|---|---|---|---|
| |
CB_reconstruction_LS_EM.gif | manage | 20 K | 2007-06-26 - 17:01 | JacekOtwinowski | S_CB |
| |
CB_reconstruction_comb_LS_EM.gif | manage | 31 K | 2007-06-26 - 17:20 | JacekOtwinowski | S_CB_3 |
| |
CB_reduction.gif | manage | 11 K | 2007-06-26 - 17:20 | JacekOtwinowski | S_CB_2 |
| |
Double_hits_epem.gif | manage | 6 K | 2007-07-05 - 02:08 | JacekOtwinowski | dh1 |
| |
Double_hits_epep.gif | manage | 6 K | 2007-07-05 - 02:09 | JacekOtwinowski | dh2 |
| |
Signal_and_CB.gif | manage | 34 K | 2007-06-27 - 17:19 | JacekOtwinowski | S_CB_1 |
Edit | Attach | Print version | History: r12 < r11 < r10 < r9 | Backlinks | View wiki text | Edit wiki text | More topic actions
Topic revision: r12 - 2007-07-05, JacekOtwinowski
Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors.
Ideas, requests, problems regarding Foswiki Send feedback | Imprint | Privacy Policy (in German)
Ideas, requests, problems regarding Foswiki Send feedback | Imprint | Privacy Policy (in German)


