- The e- and e+ identification by using HPidAlgStandCuts algorithm
- e+/e- signal eveluation in RICH
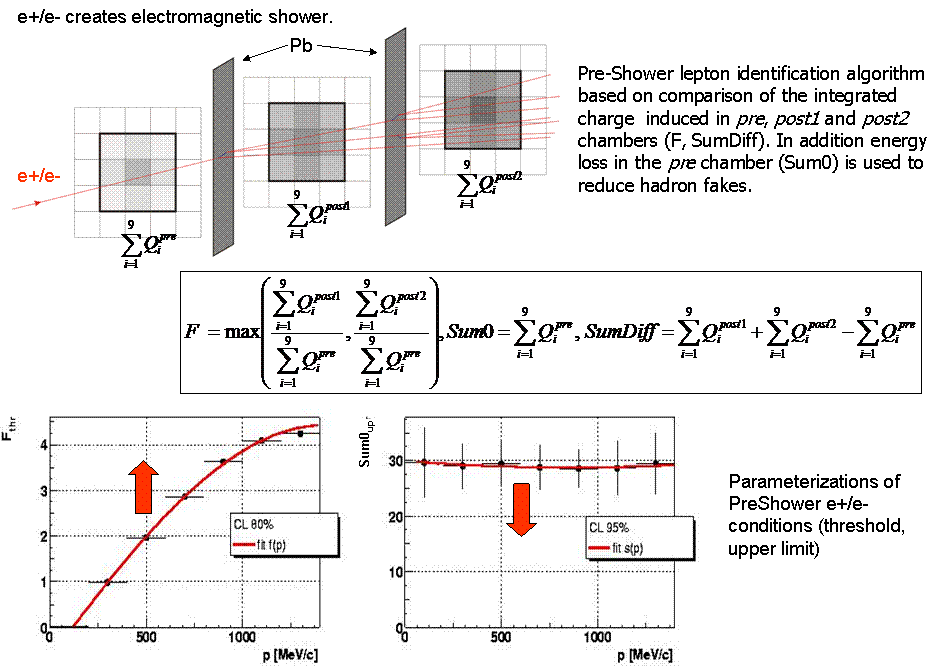
- e+/e- identification in PreShower
- HPidAlgStandCutsPar parameter container
- How to run HPidAlgStandCuts algorithm in the HYDRA framework
- Example of programs
- Exercises (HPidAlgStandCutsPar paramter container)
- Exercises (control PID ntuple with C+C@2GeV events)
The e- and e+ identification by using HPidAlgStandCuts algorithm
There are several PID algorithms implemented in the HYDRA package which are used to identify particle species. One of them isHPidAlgStandCuts algorithm which can be used to identify e+/e- from elementary and heavy ion collisions. At the moment this algorithm uses information (observables) provided by RICH (ring properties), MDC (momentum, track length), TOF/TOFino (tof) and PreShower (elctromagnetic shower properties) detectors. The RICH detector plays a crutial role in e+/e- identification in the HADES spectrometer because it is almost blind to hadrons at beam energies of 1-2 AGeV.
Thus, each electron candidate must have correlated signal in the RICH detector. It is realised by requiring the angular correlation between RICH and inner MDC segment.
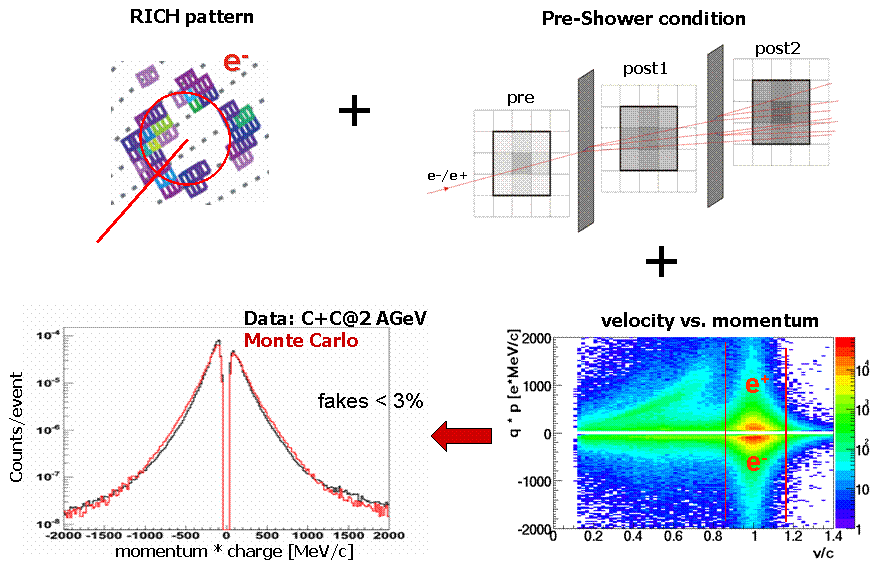
e+/e- signal eveluation in RICH
The RICH ring properties used inHPidAlgStandCuts for e+/e- identification.
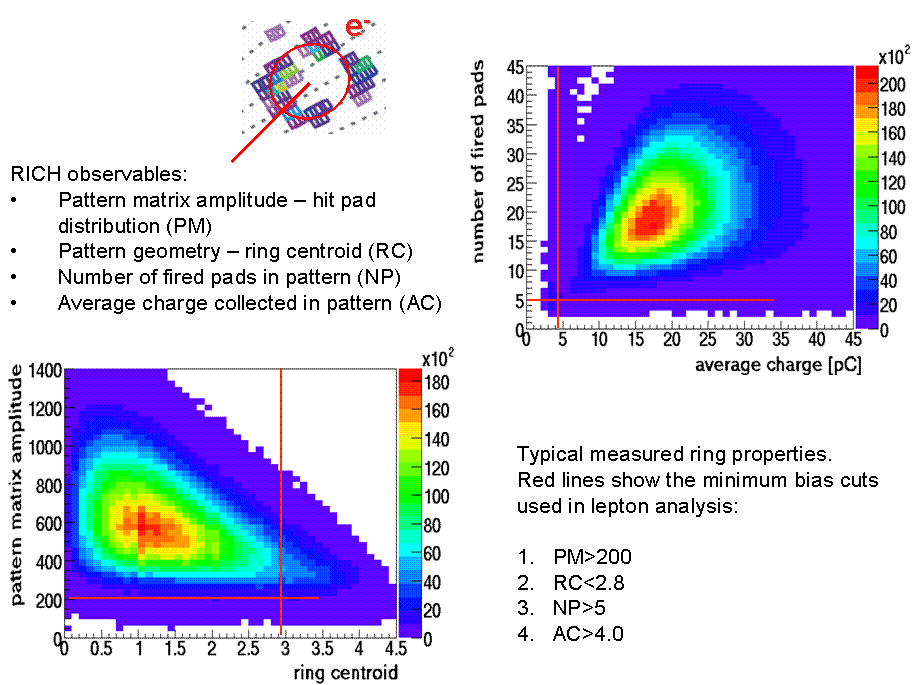
e+/e- identification in PreShower
The PreShower observables used inHPidAlgStandCuts for e+/e- identification.
HPidAlgStandCutsPar parameter container
TheHPidAlgStandCutsPar is a parameter container which is used by HPidAlgStandCuts reconstructor for e+/e- identification. It derives from HPidHistogramsCont and currently contains 7 parameter sets (histograms) for 7 different e+/e- identification algorithms. These histograms are stored in HPidAlgStandCutsPar in the following way:
HPidAlgStandCutsPar *par = rtdb->getContainer("PidAlgStandCutsPar");
par->addHistogram(particleID, algorithmOffset, sectorNb, pParamHist);
// set parameter context
par->setParamContext("HPidAlgStandCutsParContextKICK");
where particleID is particle ID (Geant convension: 2 - e+, 3 - e- ...) , algorithmOffset is an algorithm offset, sectorNb is a sector number and pParamHist is a pointer to histogram with algorithm parameters. The parameter context defines the track reconstruction algortihm. The algorithmOffset and pParamHist for HPidAlgStandCuts are following:
algorithmOffset |
pParamHist |
| 0 | Rich ring parameters |
| 1 | PreShower F_thr vs. momentum parametrization |
| 2 | PreShower Sum0_up vs momentum parametrization |
| 3 | system0 (momentum vs. beta - lower limits) |
| 4 | system0 (momentum vs. beta - upper limits) |
| 5 | system1 (momentum vs. beta - lower limits) |
| 6 | system1 (momentum vs. beta - upper limits) |
| 7 | Pre-Shower SumDiff_thr vs. momentum parametrization |
HPidAlgStandCutsPar parametrs:
- Create and fill histograms (sectorwise, particleID) with
- Rich ring properties (
PM, RC, NP, AC) -
momentum vs betafor the system0 (PreShower polar angles) and for system1 (TOF polar angles) - PreShower (
F vs momentum, Sum0 vs momentum and SumDiff vs momentum)
- Rich ring properties (
- Define and calculate parameter thresholds, upper or lower limits (e.g. Rich minimum bias cuts, momentum vs. beta (3 sigma), F_thr (CL =80%)...)
- Parametrize
F_thr(momentum),Sum0_up(momentum)andSumDiff_thr(momentum)dependencies by fitting the functions - Store histograms with algorithm parameters in the
HPidAlgStandCutsParas described above
How to run HPidAlgStandCuts algorithm in the HYDRA framework
In order to make PID by usingHPidAlgStandCuts algorithm two HYDRA reconstructors HPidReconstructor and HPidParticleFiller must be used.
The HPidReconstructor is a master reconstructor which calls algorithms (e.g. HPidAlgStandCuts algorithm) making particle identification (PID)
while the HPidParticleFiller selects identified particles and stores them in the output HPidParticle category.
There are following flags implemented in the HPidAlgStandCuts algorithm:
RICHRINGCUTS - Rich ring properties TOFPVSBETA - momentum vs. beta window (TOF) TOFINOPVSBETA - momentum vs beta lower limit (TOFINO) SHOWERMAXFVSP - threshold on charge multiplication factor max(Sum1/Sum0,Sum2/Sum0) in PreShower SHOWERSUM0VSP - upper limit on charge Sum0 in the first chamber of PreShower SHOWERSUMDIFFVSP - threshold on charge difference Sum1+Sum2-Sum0 in PreShowerUser can select conditions which should be used by
HPidAlgStandCuts for PID.
By default the "RICHRINGCUTS,TOFPVSBETA,TOFINOPVSBETA,SHOWERMAXFVSP,SHOWERSUM0VSP" flags are used in HPidAlgStandCuts algorithm.
The HPidAlgStandCuts was designed in such a way that it returns 0 (is not lepton) or 1
(is lepton) for the selected track/momentum reconstruction algorithm which has to be specified
by user in the HPidReconstructor constructor. The identification of other paricles pions/protons/deuterons is not done by this algorithm and decision is not taken.
To run PID by using HPidAlgStandCuts algorithm you need the
HPidTrackCand or HPidTrackCandSim category in the input DST file and
the set of parameters HPidAlgStandCutsPar which are initialised from ORACLE. They can be different
for different track/momentum reconstruction algorithms (KICK,SPLINE, RUNGEKUTTA) and must be stored in ORACLE
with suitable context.
The HPidAlgStandCuts algorithm creates the output control ntuple (optional)
which contains useful information about standard cuts performance.
The HPidParticleFiller task rejects particles for which HPidAlgStandCuts
returns 0. It is done for selected track/momentum reconstruction algorithm which must be
set by user. Only tracks reconstructed by this algorithm are stored in the output
HPidParticle data category.
User must select the same track/momentum reconstruction algorithm for both
HPidReconstructor and HPidParticleFiller reconstructors.
In orer to use the HPidAlgStandCuts algorithm (experiment) in the HYDRA framework the following lines of code have to be added to the analysis program:
// create HPidReconstructor reconstructor (must be added later to task set)
HPidReconstructor *pPidRec=new HPidReconstructor("pdf,ALG_RUNGEKUTTA")
Short_t nParticles[2]={2,3}; // only lepton ID
pPidRec->setParticleIds(nParticles,sizeof(nParticles)/sizeof(Short_t));
// create HPidAlgStandCuts algorithm and add it to HPidReconstructor (default cuts)
HPidAlgStandCuts *pPidAlgStandCuts=new HPidAlgStandCuts("nameOutputNtuple.root");
pPidRec->addAlgorithm(pPidAlgStandCuts);
// create HPidParticleFiller (must be added later to the task list)
HPidParticleFiller *PartFiller=new HPidParticleFiller("RUNGEKUTTA");
pPartFiller->setAlgorithm(7); // 7-check values from HPidAlgStandCuts algorithm
// add user reconstructors/tasks to the master task set
HTaskSet *pTaskSet=gHades->getTaskSet("real");
pTaskSet->add(pPidRec);
pTaskSet->add(pPartFiller);
Example of programs
executable program to run PID by usingHPidAlgStanCuts algorithm
Exercises (HPidAlgStandCutsPar paramter container)
- Initialise
HPidAlgStandCutsParparameter container from ORACLE and store it in ROOT file (runID=7006)- Set your HYDRA environement by using
defall.sh - Copy
/d/hades/jacek/hydra-8.13/tutorials/initParameters/initPidFromOra.Cmacro to your working directory - Open the macro and identify:
paramater container,runIdandoutput file - Run the macro (
root -l initPidFromOra.C)
- Set your HYDRA environement by using
- Draw parameter histograms stored in
HPidAlgStandCutsPar- Open the
output filefrom produced byinitPidFromOra.C - Draw parameter histograms by using
PidAlgStandCutsPar->getHistogram(particleID,algorithmOffset,sectorNb)->Draw()function
- Open the
Exercises (control PID ntuple with C+C@2GeV events)
- Open wiki page with ntuple variable description (http://hades-wiki.gsi.de/cgi-bin/view/Computing/HPidAlgStandCutsControlNtuple)
- Make q*p vs. cut number plots
- Open
/d/hades06/data/sudol/nov02/gen4/leptons/new/leptons_gen4.rootroot ntuple - Create and draw q*p histograms vs. cuts numbers:
-
pLep->Draw("charge*mom>>h0(201,-1000,1000)") -
pLep->Draw("charge*mom>>h1(201,-1000,1000)","isrich") -
pLep->Draw("charge*mom>>h2(201,-1000,1000)","isrich&&(istof||istofino)") -
pLep->Draw("charge*mom>>h3(201,-1000,1000)","isrich&&(istof||istofino&&isshower)")
-
- Draw all histograms in one ROOT canvas (e.g.
h0->Draw(); h1->Draw("same");)
- Open
- Determine purity and efficiency of e+/e- standard cuts reconstruction algorithm
- Open
/d/hades06/data/sudol/nov02/gen4/urqmd/leptons_urqmd_gen4.rootsimulation ntuple - Draw
gcomtrkntuple variablepLep->Draw("gcomtrk")//piddef.h file //Enumerated variables indicating which detector has seen a geant particle //and combinations thereof (to be used as shorthand-notation). These are gcomtrk values enum enumDetBits { //individual detectors kTrackNotSet = 0x00, // This object does not contain valid data kSeenInRICHIPU = 0x01, // RICH IPU has seen this trackid kSeenInOuterMDC = 0x02, // Outer Mdc has seen this trackid kSeenInRICH = 0x04, // RICH has seen this trackid kSeenInInnerMDC = 0x08, // Inner Mdc has seen this trackid kSeenInTOF = 0x10, // TOF has seen this trackid kSeenInShower = 0x20, // Shower has seen this trackid kSeenInMETA = 0x40, // One of the Meta dets has seen this trackid }; - Create and draw q*p histograms before cuts (only pure tracks)
-
pLep->Draw("charge*mom>>h0(201,-1000,1000)","gcomtrk>74")
-
- Create and draw q*p histograms after all cuts (only pure tracks)
-
pLep->Draw("charge*mom>>h1(201,-1000,1000)","isrich&&(istof||istofino&&isshower)&&gcomtrk>74")
-
- Create and draw q*p histograms after all cuts
-
pLep->Draw("charge*mom>>h2(201,-1000,1000)","isrich&&(istof||istofino&&isshower)")
-
- Efficiency is difined as (
Eff = h1/h0) while Purity is defined as (Pur = h1/h2)- Use ROOT
Clone()function to create clone ofh0,h1andh2histograms (e.g.TH1F* h0_clone=h0->Clone()) - Use ROOT
Sumw2()function forh1andh1_clonehistograms to propagate properly errors (e.g.h1->Sumw2()) - Calculate efficiency (
h1->Divide(h0_clone)) and purity (h1_clone->Divide(h2_clone)) - Draw efficiency (
h1->Draw()) and purity (h1_clone->Draw())
- Use ROOT
- Open
| I | Attachment | Action | Size | Date | Who | Comment |
|---|---|---|---|---|---|---|
| |
ElectronID.gif | manage | 55 K | 2007-06-26 - 14:41 | JacekOtwinowski | ElectronID |
| |
PreShowerElectronID.gif | manage | 41 K | 2007-06-25 - 13:44 | JacekOtwinowski | PreShowerElectronID |
| |
RichElectronID.gif | manage | 61 K | 2007-06-25 - 13:43 | JacekOtwinowski | RichElectronID |
This topic: Homepages > HomepageJacekOtwinowski > TeachingWorkshop > PID-PidAlgStandCuts
Topic revision: 2007-07-03, JacekOtwinowski
Topic revision: 2007-07-03, JacekOtwinowski
Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors.
Ideas, requests, problems regarding Foswiki Send feedback | Imprint | Privacy Policy (in German)
Ideas, requests, problems regarding Foswiki Send feedback | Imprint | Privacy Policy (in German)

